GRAM-POSITIVE COCCI
LEARNING OBJECTIVES
Perform the biochemical tests used to identify Staphylococcus
Differentiate Staphylococcus from other gram-positive cocci found in clusters on Gram stain
Perform biochemical tests used to identify Streptococcus and Enterococcus
Differentiate pathogenic Streptococcus and Enterococcus
MCCCD OFFICIAL COURSE COMPETENCIES
Describe the modes of bacterial and viral reproduction and proliferation
Utilize aseptic technique for safe handling of microorganisms
Apply various laboratory techniques to identify types of microorganisms
Identify structural characteristics of the major groups of microorganisms
Compare and contrast prokaryotic cell and eukaryotic cell
Compare and contrast the physiology and biochemistry of the various groups of microorganisms
MATERIALS
Students will complete the lab in groups of 3 at Pecos and groups of 4 at Williams
Cultures: Please pick up 1 of each of the numbered (1-7) gram-positive cocci cultures.
Staphylococcus aureus slant culture
Staphylococcus epidermidis slant culture
Micrococcus luteus slant culture
Micrococcus roseus slant culture
Group C Streptococcus slant culture
Streptococcus sanguinis slant culture
Enterococcus faecalis slant culture
Media:
Students will complete the lab in groups of 3 at Pecos and groups of 4 at Williams
7 glucose fermentation tubes per group
4 Mannitol Salt Agar plates per group
4 blood agar plates per group
7 bile esculin slant per group
Equipment:
Bottle of hydrogen peroxide
Microscope slides
Inoculating loops
Test tube rack
Bacticinerators
BIOCHEMICAL TESTS ALBUM LINK
The gram-positive cocci and gram-negative bacilli lab exercises will allow you to gain the knowledge and skills you need to complete the capstone project in BIO205, the mixed unknown. In the mixed unknown project you will use biochemical testing to identify one gram-positive cocci and one gram-negative bacilli. This lab exercise focuses on gram-positive cocci.
Gram-positive cocci are ubiquitous in nature. When gram-positive cocci are isolated from a clinical specimen, the first consideration is to determine if the isolate is a member of the Micrococcaceae (Staphylococcus or Micrococcus) or the Streptococcaceae family (Streptococcus or Enterococcus). In this exercise, you will learn how to identify four members of the Micrococcaceae family (Staphylococcus aureus, Staphylococcus epidermidis, Micrococcus luteus, and Micrococcus roseus) and three members of the Streptococcaceae family (Streptococcus sanguinis, Group C Streptococcus, and Enterococcus faecalis). These gram-positive cocci are normally found on the skin and mucous membranes in the mouth, nasal passages, throat, anus and vagina.
Gram stains and other differential stains can help us group bacteria, but do not identify the bacteria according to their genus and species. In a Gram stain Staphylococcus or Micrococcus are often seen in grape-like clusters, Streptococcus and Enterococcus are seen in pairs or chains.
Colony characteristics can aid in the identification of bacteria. Colonies of Staphylococcus or Micrococcus are usually larger (2 to 3 mm after 24 hours of incubation) than Streptococcus or Enterococcus colonies (pinpoint or smaller than 2 mm after 24 hours of incubation). Staphylococcus and Micrococcus colonies are usually convex, opaque and frequently pigmented.
How are bacteria identified using biochemical testing? Usually, several biochemical tests are performed to establish unique patterns of growth and metabolic processes to identify bacteria. Biochemical testing includes testing for enzymes and the use of selective and differential media. Bacteria metabolize protein, carbohydrates, and lipids to produce energy in much the same way that humans do. Their ability to metabolize these macromolecules is dependent on their ability to produce the specific enzymes that catalyze the biochemical reactions. Since enzymes are proteins, a bacterium’s DNA will determine what enzymes an organism can produce. Since DNA is not the same in all bacteria, bacteria will differ in their ability to produce specific enzymes.
When a bacteria produces an enzyme and a metabolic biochemical reaction occurs, end products will be produced. We can test for the presence of enzymes by studying these biochemical processes and detecting the products of the reactions. For example, Staphylococcus and Micrococcus produce the catalase enzyme, Streptococcus and Enterococcus do not produce catalase. Often the products of a biochemical reaction will produce a change in pH of the media in which they occur. If we add a pH indicator to the media, we often can tell if a biochemical reaction occurs by a change in the color of the media. Other times a biochemical reaction will destroy one of the components that makes up the media and the media will physically change in some way. Hemolysis (destruction of red cells) is an example of a change in media composition because of microbial enzymes.
Selective media contains substances that allow some microorganisms to grow but prevent the growth of other organisms. Differential media contains substances that help distinguish microorganisms that grow on the media either by changes that occur in the media or by changes that occur in the appearance of the microorganism. Selective and/or differential media may be used in Petri plates or in tube media (both solid and liquid).
Staphylococcus aureus is usually considered to be pathogenic, but it may be found in the absence of disease. Some people are carriers of this organism, meaning they harbor the pathogen but exhibit no symptoms. One study found as many as 90% of hospital personnel were S. aureus carriers! This poses a risk for hospitalized patients whose immune systems may already be stressed or compromised by their underlying condition. MRSA is especially dangerous because these isolates have become resistant to penicillin derivatives, such as methicillin and oxacillin.
Staphylococcus aureus produces enzymes (hyaluronidase and collagenase) to help it invade and destroy healthy tissue. Staphylococcus aureus produces coagulase and staphylokinase that hijack the host’s coagulation system. It can cause infections ranging from superficial (boils, impetigo) to systemic infections which involve deeper tissues, such as bone (osteomyelitis), lung (pneumonia), heart valves (endocarditis), brain (meningitis), and blood (toxic shock syndrome). Staphylococcus aureus is the most common etiologic agent of wound infections, including surgical infections. It is a common cause of Healthcare Acquired/Associated Infections (HAI) which are acquired in hospitals and other healthcare facilities. And, many strains produce an exotoxin which causes food poisoning.
Staphylococcus epidermidis is the predominant species of Staphylococcus found on the skin and usually not pathogenic. Micrococcus luteus and Micrococcus roseus are also not pathogenic, though it is important to remember that under the proper circumstances, almost any bacteria can cause disease.
The Streptococcus can be classified according to their hemolytic properties. After determining the hemolysis, subsequent biochemical testing can narrow down the identification even further.
Some bacteria produce hemolysins that damage the membrane of red blood cells. This process is called hemolysis. The degree to which the blood cells are hemolyzed is used to distinguish bacteria from one another. There are three types of hemolysis, designated as beta, alpha and gamma.
Beta hemolysis represents a complete lysing of the red blood cells. This is demonstrated by a complete clearing of the red blood cells in the media around a colony.
Alpha hemolysis represents a partial damage of the red blood cells which reduces the hemoglobin to methemoglobin. This partial damage is shown as an olive green to brown discoloration in the media surrounding the bacterial colony on the blood agar.
Gamma hemolysis represents the lack of hemolysis. There is no damage to the red blood cells in the media.
Group C streptococci are a common cause of infection in several animal species but are generally considered to be a rare cause of infection in humans.
Streptococcus sanguinis is considered part of the normal microbiome in the mouth and forms oral biofilms known as dental plaque. It also is the most common streptococcus associated with bacterial endocarditis (infection of the heart valve or endocardium). Streptococcus mutans is another streptococcus associated with dental caries and plaque.
The enterococci are among the most ubiquitous bacteria known. They are found in human and animal gastrointestinal tracts and the environment. There are many different species of enterococci, but only a few have the potential to cause infections in humans. More than 95% of infections due to enterococci are caused by just two species, Enterococcus faecium and Enterococcus faecalis. These bacteria are not very invasive but are significant opportunists causing urinary tract infections, pelvic abscesses, peritonitis and wound infections in humans. Recently many strains have acquired resistance to Vancomycin and have been named “VRE” for Vancomycin Resistant Enterococcus.
PRE-ASSESSMENT
PROCEDURE
For this exercise: Work in groups of 3 at Pecos groups of 4 at Williams. Perform the following tests on each gram-positive cocci numbered culture.
CATALASE TEST
This test determines the ability of the bacterium to produce the enzyme catalase. Hydrogen peroxide is toxic to the cell when it accumulates during aerobic respiration. The bacteria must be able to break down H2O2 into H2O and O2 to survive.
This is first test used to differentiate Staphylococcus and Micrococcus from Streptococcus and Enterococcus.
Step 1. Obtain a glass slide and a small bottle of hydrogen peroxide.
Step 2. Using a sterilized inoculating loop, smear a small amount of bacteria onto the dry slide. Place the slide on a dark background for better observation.
Step 3. Place a drop of hydrogen peroxide on top of the bacterial smear. The immediate appearance of bubbles indicates the organism can produce catalase.
Bubbles = Positive
No bubbles = Negative
Step 4. Record the result on the worksheet.
Step 5. Dispose of the slide in the used slide basin.

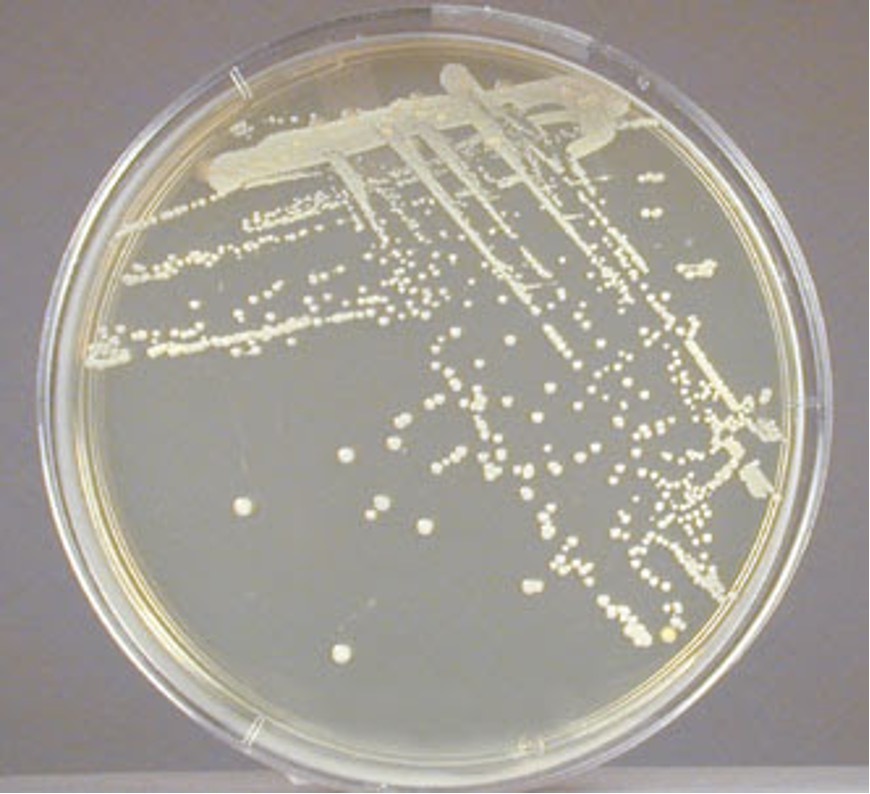
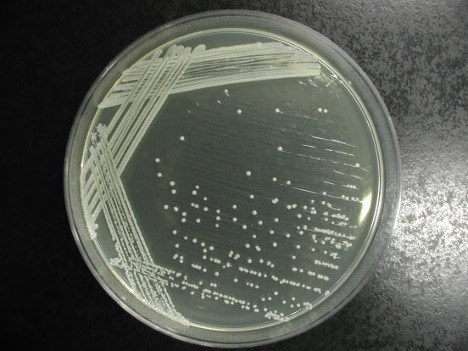
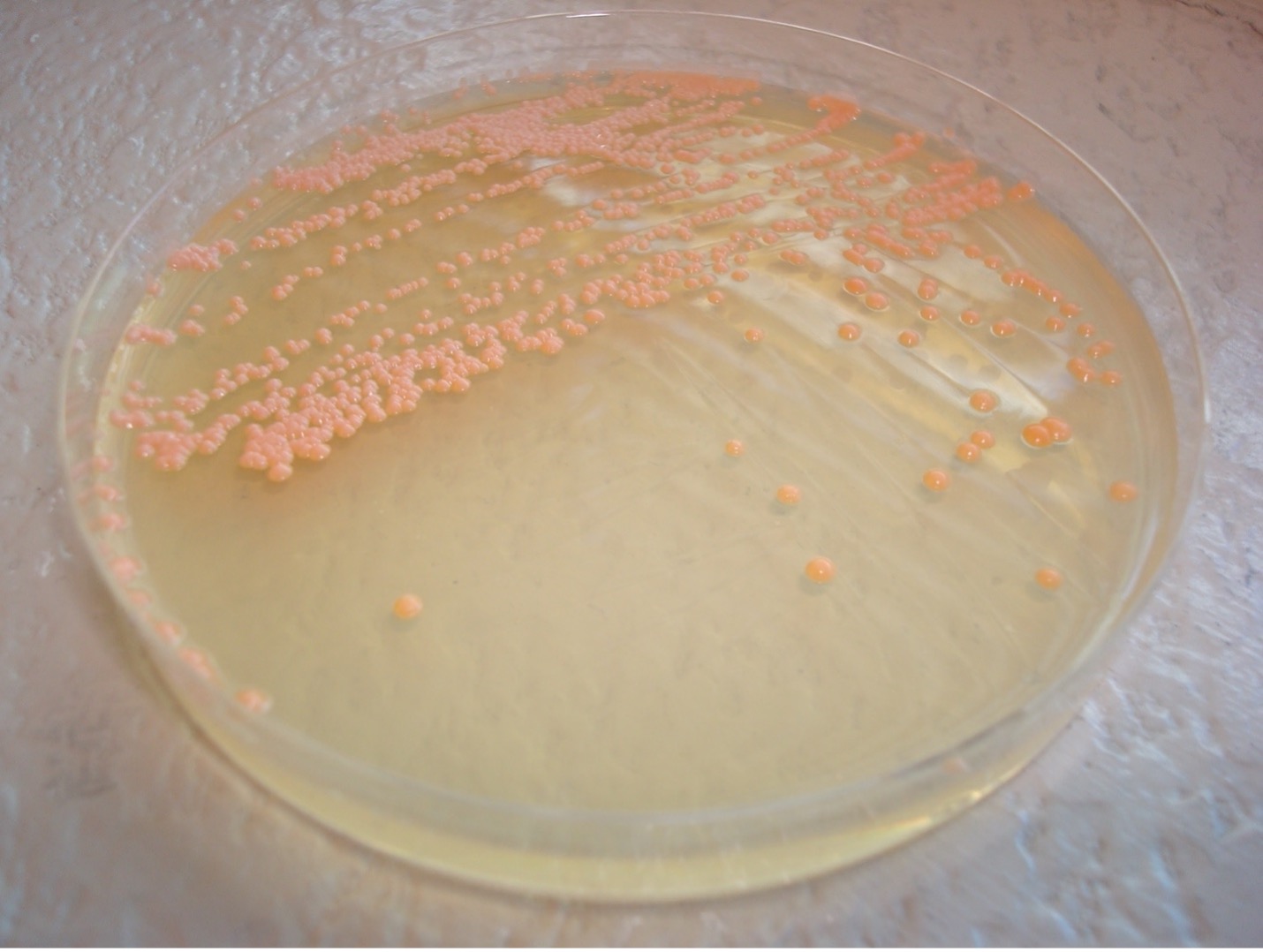
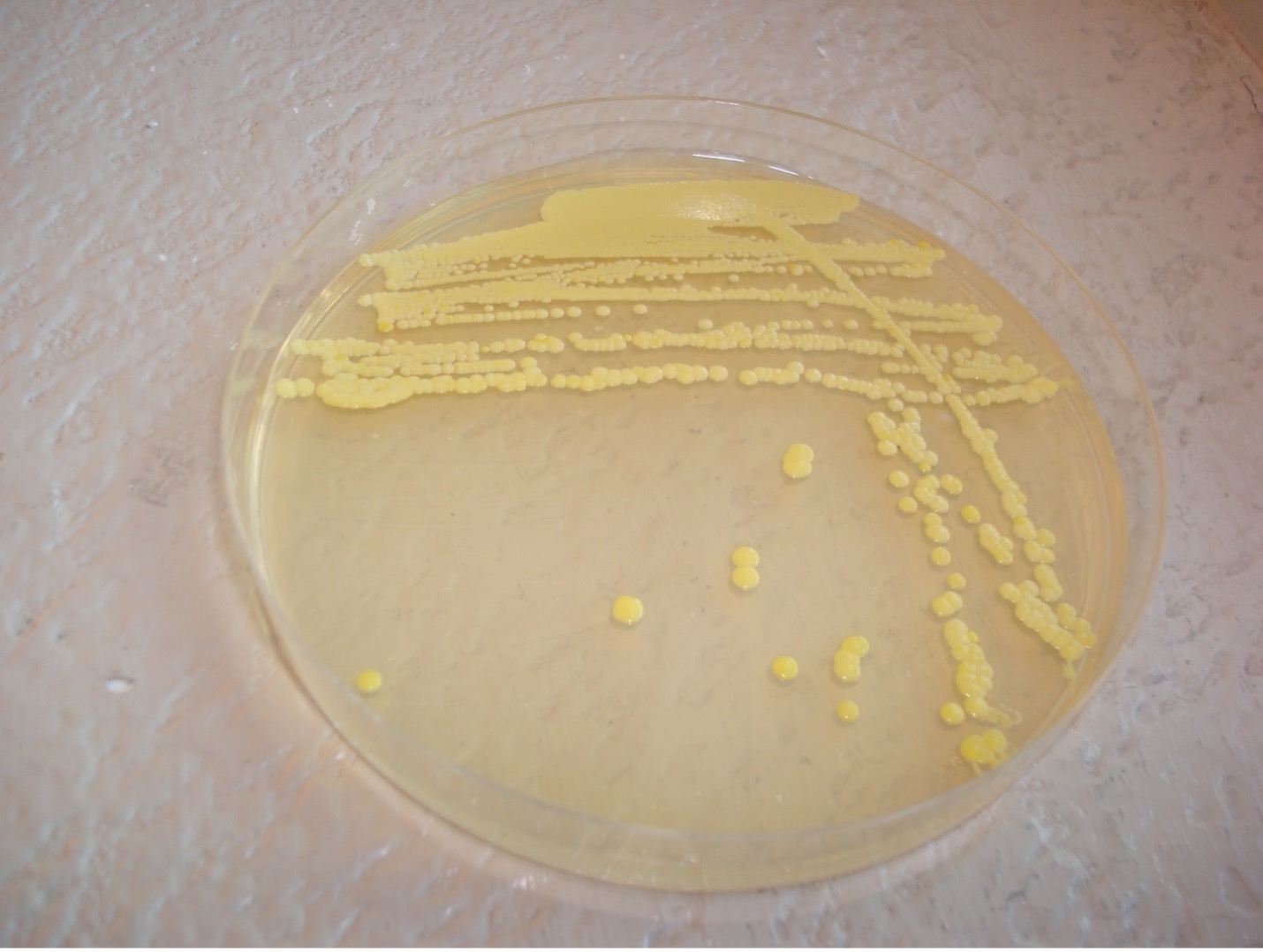
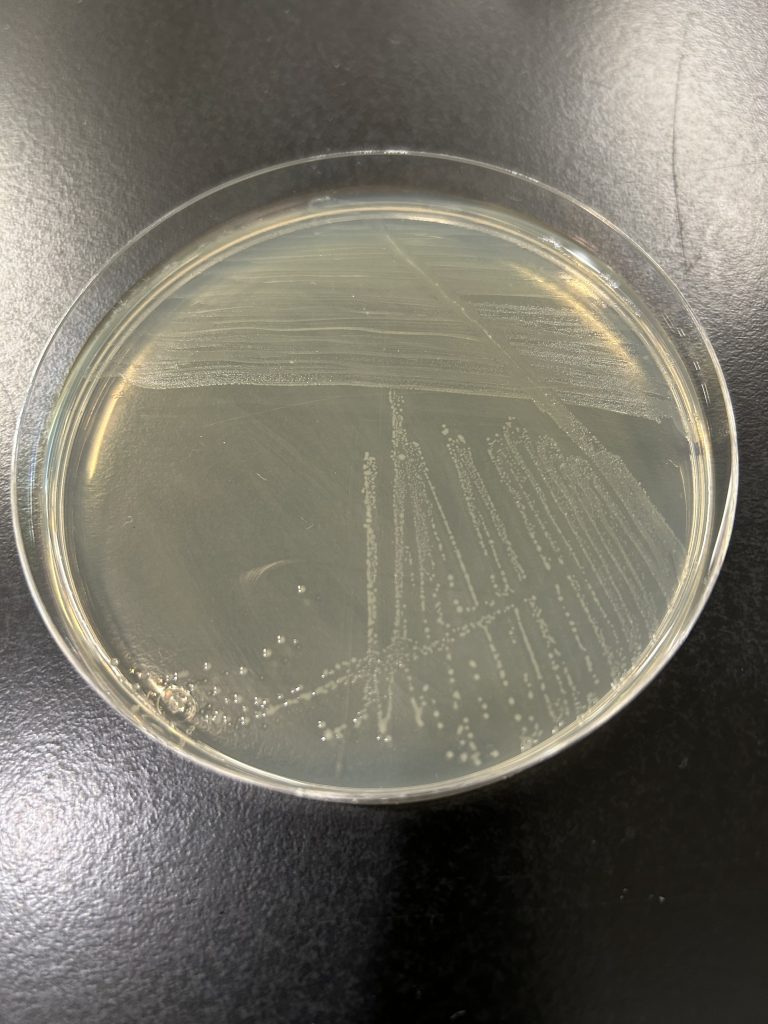
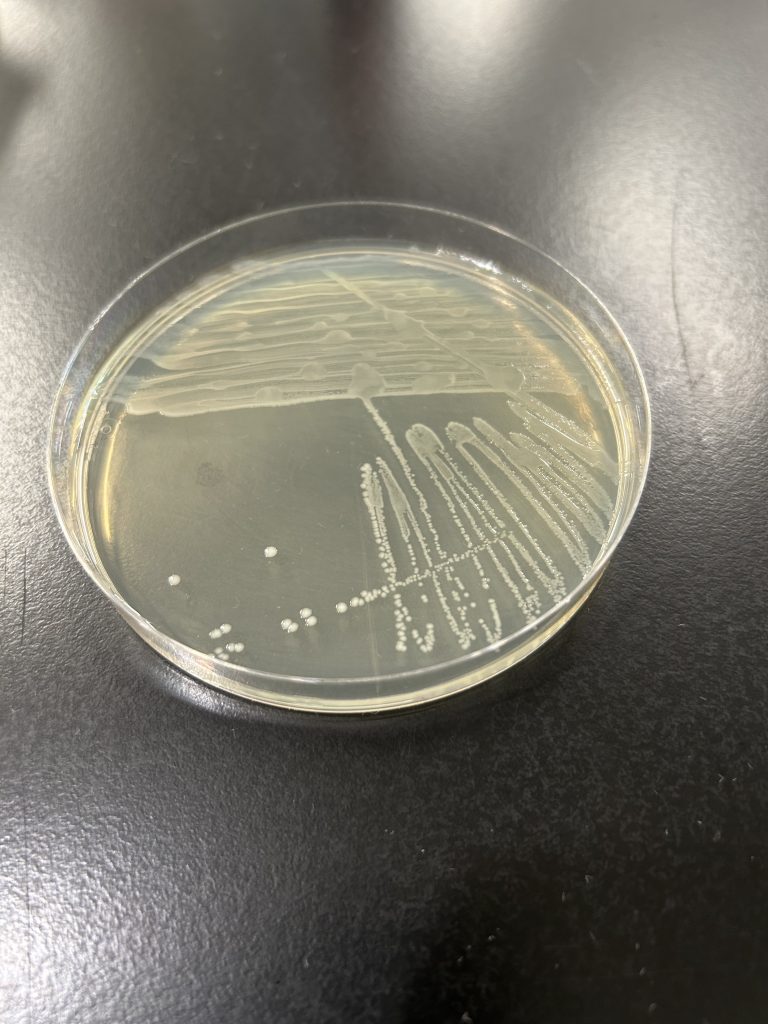
colony color
Staphylococcus colonies are white while some species of Micrococcus will produce colorful colonies (rose-coral, yellow) which can serve as an identifying characteristic. Streptococcus and Enterococcus colonies are translucent (see-through) or semi-transparent.
Step 1. Observe the growth on the TSA slant (or your gram-positive stock plate for the mixed unknown) for colony color.
Step 2. Record the color on the worksheet.
GLUCOSE FERMENTATION TEST
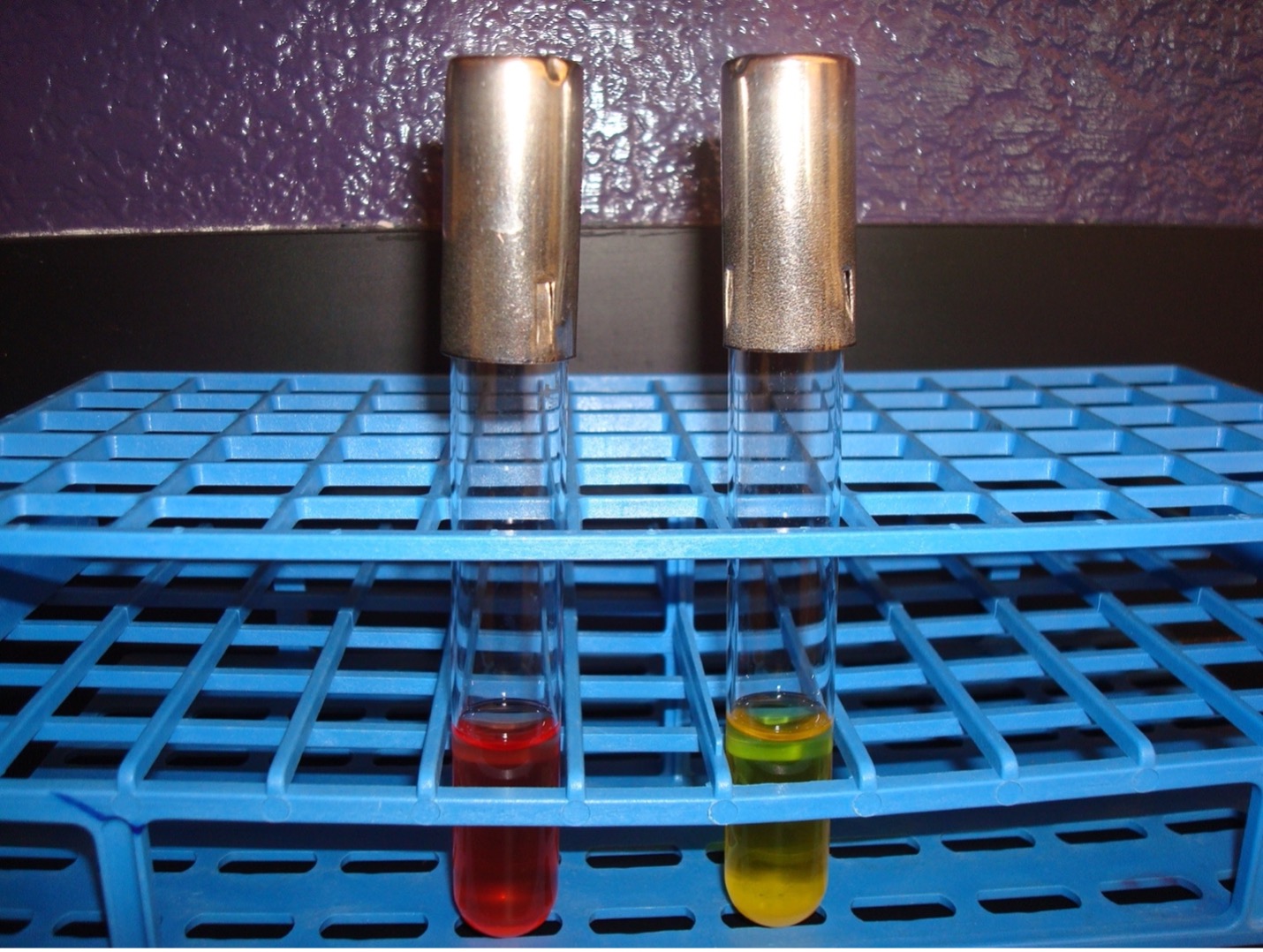
This test detects the ability of the bacterium to break down glucose to pyruvic acid. If acid is produced, the pH indicator will turn yellow.
Step 1. Obtain a glucose fermentation broth. Use a permanent marker to write the unknown number, your name (or initials), and the name of the media (glucose) on the side of the tube.
Step 2. Using a sterile inoculating loop, obtain a small amount of inoculum and inoculate the glucose fermentation broth.
Step 3. Incubate the glucose fermentation broth in the class test tube rack until the next lab session.
AFTER INCUBATION-Observe the appearance of the glucose fermentation broth. Record the result on the worksheet. Dispose of the glucose fermentation broth in the discard rack.
Yellow broth = Positive test
Red broth = Negative test
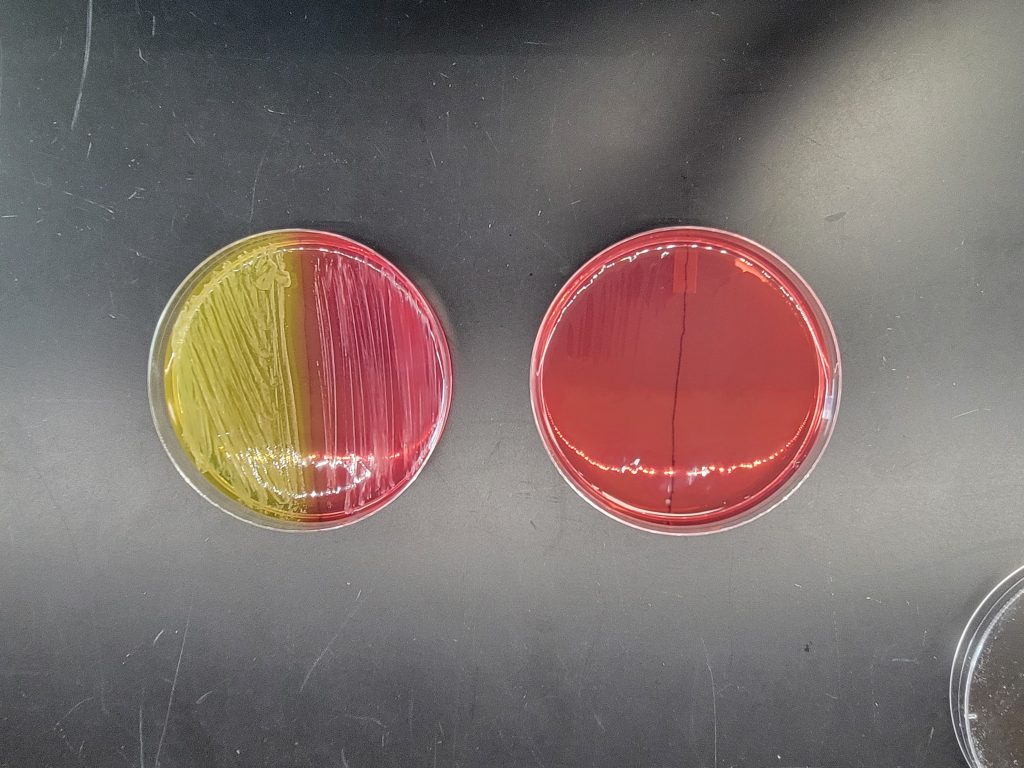
MANNITOL SALT AGAR
This is a selective and differential media which grows and differentiates Staphylococcus species. Since Staphylococcus is salt-tolerant, it will be able to grow in 7.5% salt and Staphylococcus aureus will be differentiated by mannitol fermentation.
Step 1. Each person will use one-half of a mannitol salt agar plate. Label one-half of the agar with the unknown number, your name, and the name of the media (MSA).
Step 2. Inoculate the entire side of the plate with the bacteria.
Step 3. Incubate the mannitol agar plate in the class plate tray at 37oC.
AFTER INCUBATION– There are two results to observe. Record results on the worksheet. Dispose of the mannitol salt agar plate in the biohazard trash.
Evaluate how the well the organism tolerated the salt in the agar.
Bacteria grew very well = Positive
Bacteria were inhibited from growing = Negative
If the bacteria grew very well on the media, then evaluate the organism’s ability to ferment the mannitol in the agar.
Media turned yellow = Positive = mannitol was fermented (do not confuse the yellow pigmented colonies of Micrococcus luteus as mannitol fermentation)
Media stayed reddish pink = Negative = mannitol was NOT fermented
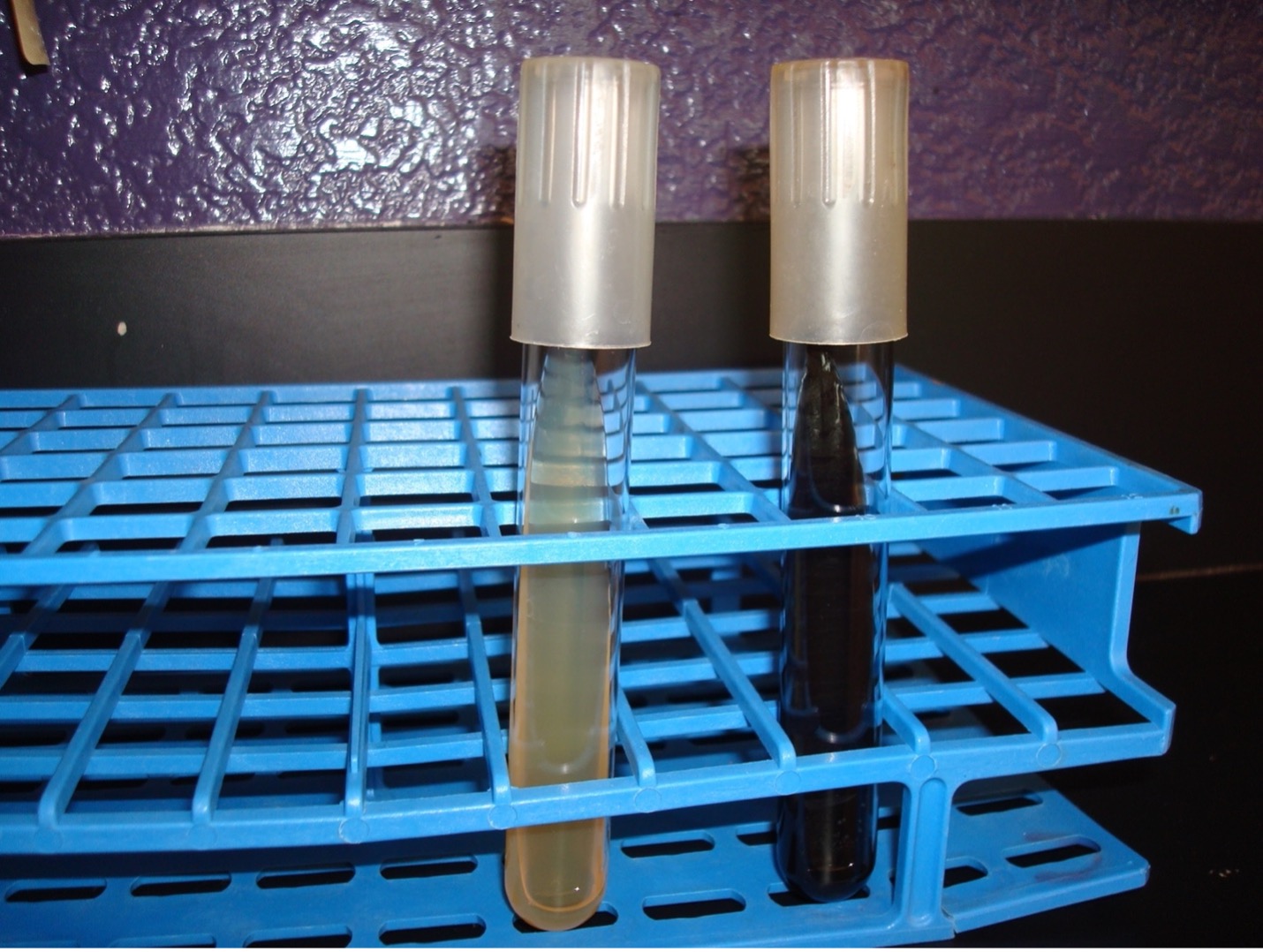
BILE ESCULIN HYDROLYSIS TEST
This test will detect the ability of the bacterium to hydrolyze bile esculin. Enterococcus faecalis can be differentiated by this test.
Step 1. Obtain one bile esculin agar slant. Use a permanent marker to write the unknown number, your name (or initials), and the name of the media (bile esculin) on the side of the tube.
Step 2. Using a sterilized inoculating loop, use a large amount of bacteria to inoculate the entire surface of the bile esculin agar slant.
Step 3. Incubate the slant in the class test tube rack until next lab session.
AFTER INCUBATION-Determine if the bacteria could hydrolyze the esculin in the agar. Record the result on the worksheet. Dispose of the Bile Esculin slant in the discard rack.
Very dark brown/black media = Positive
No color change in the media = Negative
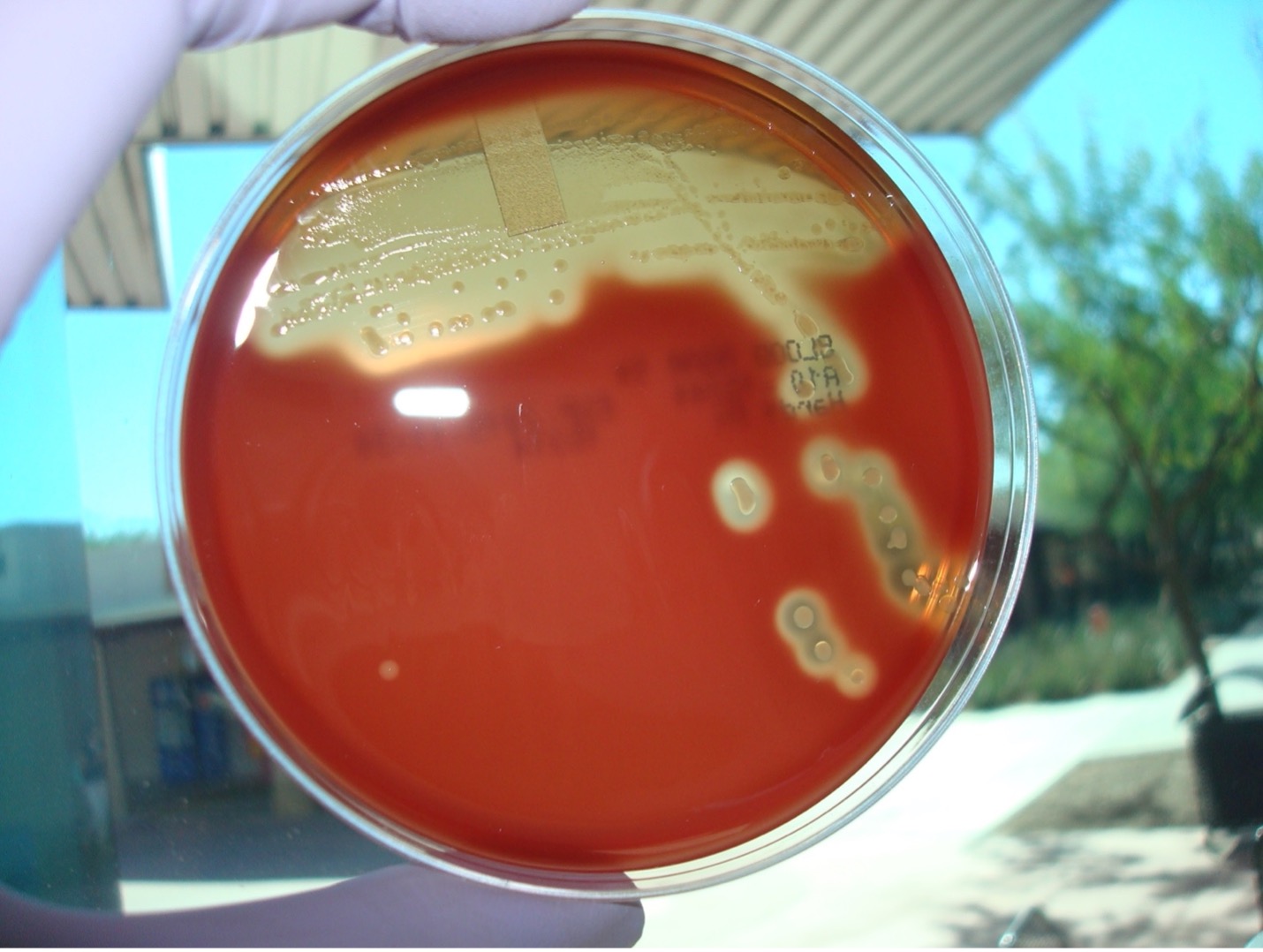
HEMOLYSIS ON BLOOD AGAR
Each person will use one-half of a blood agar plate. Label one-half of the agar with the unknown number, your name, and the name of the media (BAP).
Step 1. Inoculate the entire side of the plate with the bacteria.

Step 2. Sterilize the loop in the bacticinerator and allow it to cool.
Step 3. Stab the loop 2–3 times into the agar. Stabbing into the media will enhance the appearance of hemolysis.
Step 4. Incubate the inverted blood agar plate in the candle jar at 37oC until the next lab session. Most Streptococci grow best in a microaerophilic environment.
AFTER INCUBATION-Evaluate the type of hemolysis produced by the organisms. Hold the plate so that light is shining through the back of the agar. Determine the extent of damage to the blood in the agar surrounding the bacterial colonies. If necessary, use the inoculating loop to scrape away a few colonies to see the media underneath the colonies. Record the result on the worksheet. Dispose of the blood agar plate in the biohazard trash.
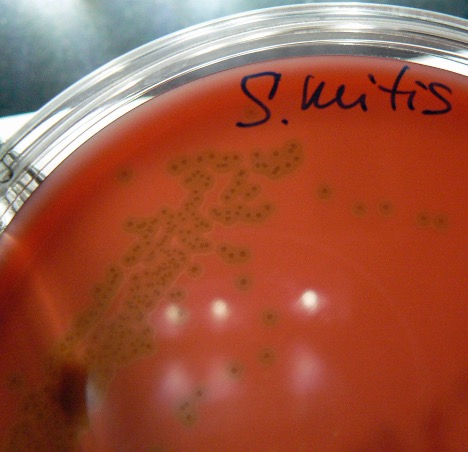
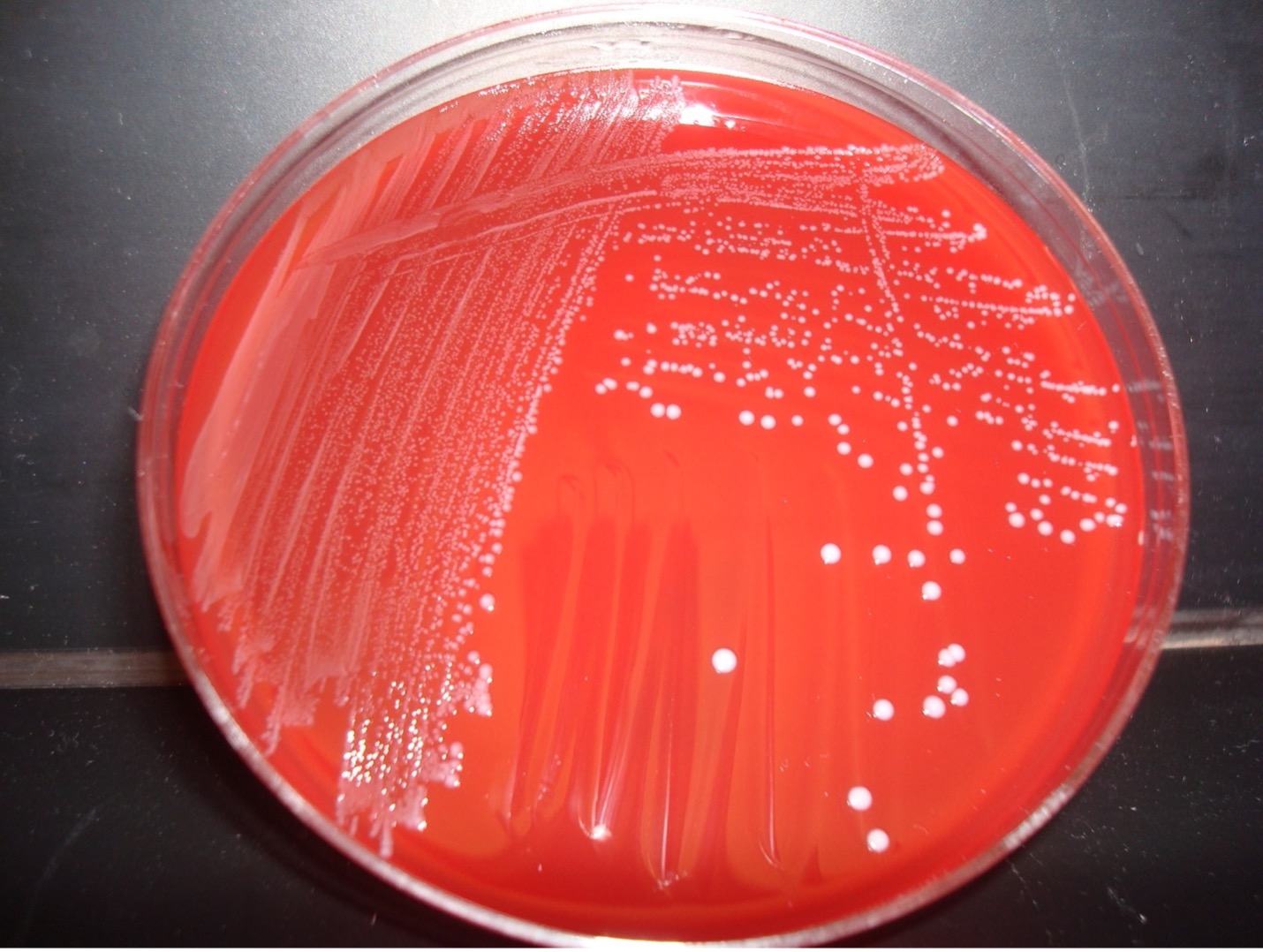
GRAM-POSITIVE COCCI BIOCHEMICAL TEST RESULTS
| TEST | Staphylococcus aureus | Staphylococcus epidermidis | Micrococcus roseus | Micrococcus luteus | Streptococcus sanguinis | Group C Streptococcus |
Enterococcus faecalis |
|---|---|---|---|---|---|---|---|
| Catalase | Positive | Positive | Positive | Positive | Negative | Negative | Negative |
| Glucose Fermentation | Positive | Positive | Negative | Negative | Positive | Positive | Positive |
| Salt Tolerance on Mannitol Salt Tolerance | Positive | Positive | Slight Positive | Positive | Negative | Negative | Positive |
| Mannitol Fermentation on Mannitol salt Agar | Positive | Negative | Negative | Negative | Negative | Negative | Positive |
| Colony color | White | White | Rose | Yellow | Translucent or semi-transparent | Translucent or semi-transparent | Translucent or semi-transparent |
| Bile Esculin | Negative | Negative | Negative | Negative | Negative | Negative | Positive |
| Hemolysis | Beta | Gamma | Gamma | Gamma | Alpha | Beta | *Variable |
NOTE: “Variable” test result means the result is not consistent. Some strains of the species will exhibit gamma hemolysis whereas other strains will produce alpha or beta hemolysis.
POST TEST
DISCOVERIES IN MICROBIOLOGY
 SARA LITTLE TURNBULL
SARA LITTLE TURNBULL
American product designer Sara Little Turnbull consulted with companies including General Mills, Coca-Cola, and Revlon. Her work with 3M would change medical care. 3M hired her to explore uses (including gift ribbons and bra cups) for a new melded polymer fiber material. At the time, she was taking care of sick family members and spending a lot of time in hospitals. She wondered if the material might block disease particles. In 1961, 3M started selling a mask based on Sara’s design. The mask blocked dust and airborne toxins, but did not block pathogens. Her mask design was used by others to create N95 masks, which are critical to stopping disease transmission.

