QUANTITATION OF MICROORGANISMS DEMONSTRATION LAB
LEARNING OBJECTIVES
Discuss viral growth requirements.
Demonstrate and recognize viral plaques.
Perform standard dilutions.
Calculate the number of organisms in a diluted sample.
MCCCD OFFICIAL COURSE COMPETENCIES
Compare and contrast viruses and cells.
Describe the modes of bacterial and viral reproduction and proliferation.
Utilize aseptic technique for safe handling of microorganisms.
Apply various laboratory techniques to identify types of microorganisms.
Identify structural characteristics of the major groups of microorganisms.
Compare and contrast prokaryotic cell and eukaryotic cell.
Compare and contrast the physiology and biochemistry of the various groups of microorganisms.
THIS IS A DEMONSTRATION LAB. PLEASE READ THROUGH THE LAB. CALCULATE THE NUMBER OF PFU/ML IN THE ONE COUNTABLE PLATE (30-300 PLAQUES) IN THE DEMONSTRATION PLATE SET PICTURED AT THE END OF THE LAB EXERCISE. RECORD YOUR CALCULATION IN THE QUANTITATION OF MICROORGANISMS DEMONSTRATION QUESTIONS DOCUMENT.
In both medical and environmental research, it is often important to know the number of microorganisms in a given sample. Some examples include:
- Determining if water from a specific source is safe to drink
- Determining if a local body of water has a low enough bacterial count to make it safe to swim in
- Determining if a food item (such as milk) is still safe to consume after its expiration date
- Determining the total number of viruses in a sample
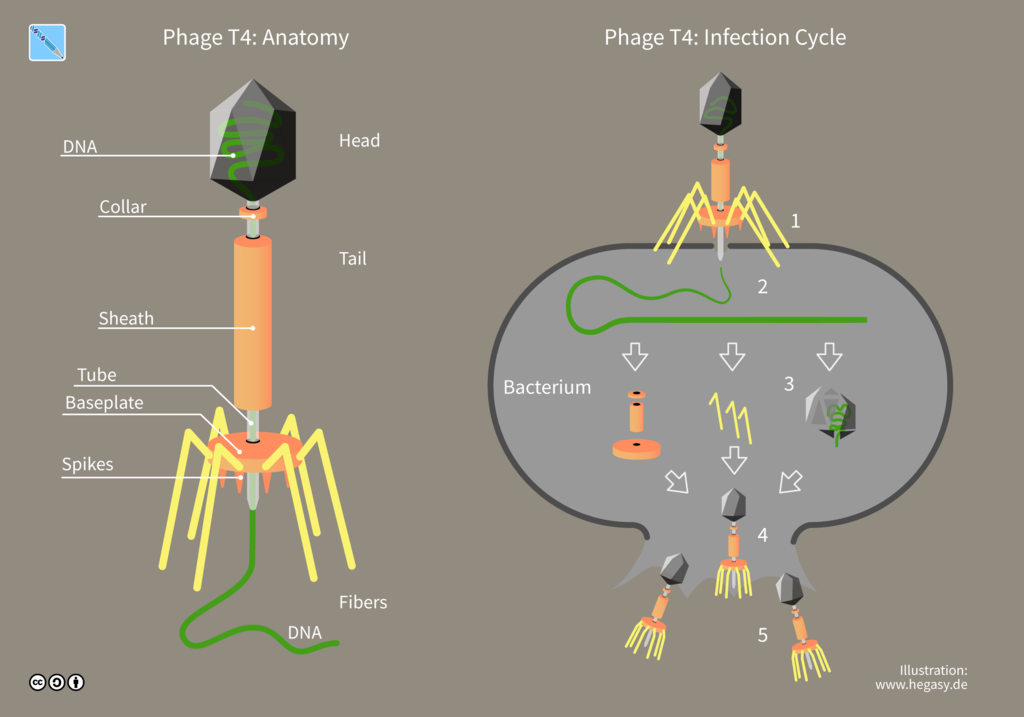
Viruses are obligate intracellular parasites, meaning they can only replicate inside another living host cell. Therefore, ordinary agar or broth media suitable for growing bacterial cultures will not sustain viral growth. However, bacteria growing in lab media can serve as the host cell for growing viruses that infect bacteria. In this lab, you will determine the total number of viruses in a given sample. You cannot count viruses, they are not visible without an electron microscope, and they reproduce inside a host cell.
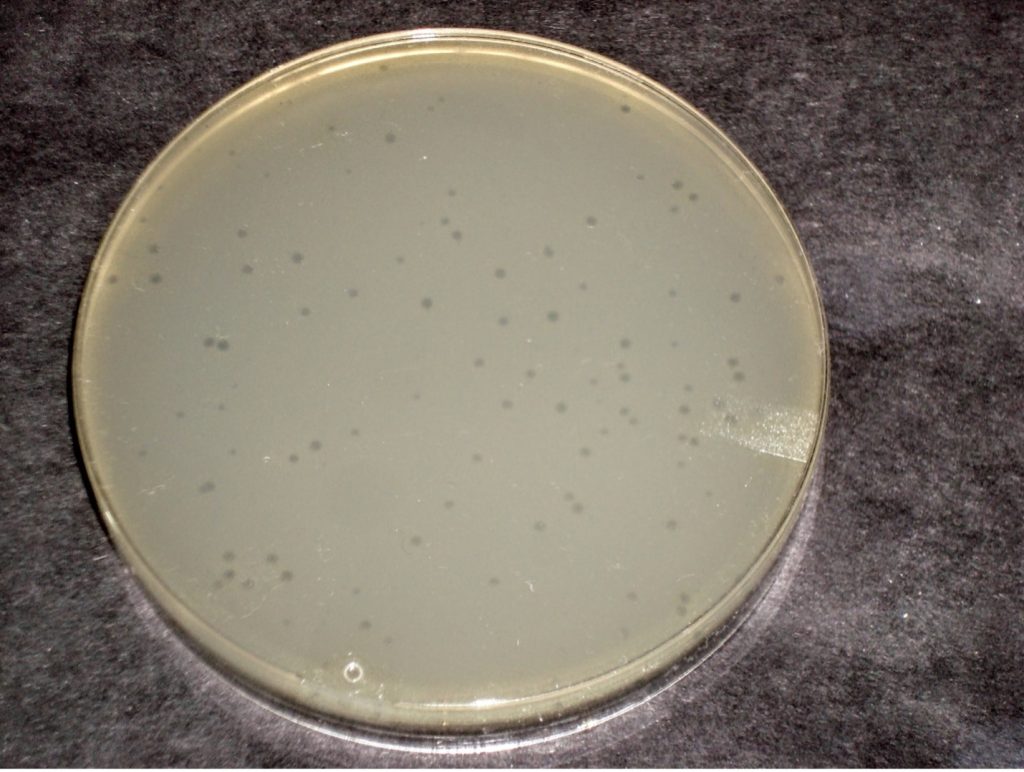
A virus that infects bacteria is called a bacteriophage or phage. A phage parasitizes bacteria and utilizes bacterial cell components to produce progeny virus. Lytic viruses burst open and kill their bacterial host once the appropriate number of viruses has been synthesized. A plaque is an area of clearing in a confluent lawn of bacterial growth which represents the spot where a virus has landed, infected the bacteria it encountered, and lysed them. Each plaque represents a virus particle or plaque-forming unit (PFU). Since there are too many bacteriophage in the original sample, it is best to dilute the sample to obtain a countable plate in which 30-300 individual plaques can be seen. We will be using the T4 bacteriophage and T4 host cells, E. coli strain B.
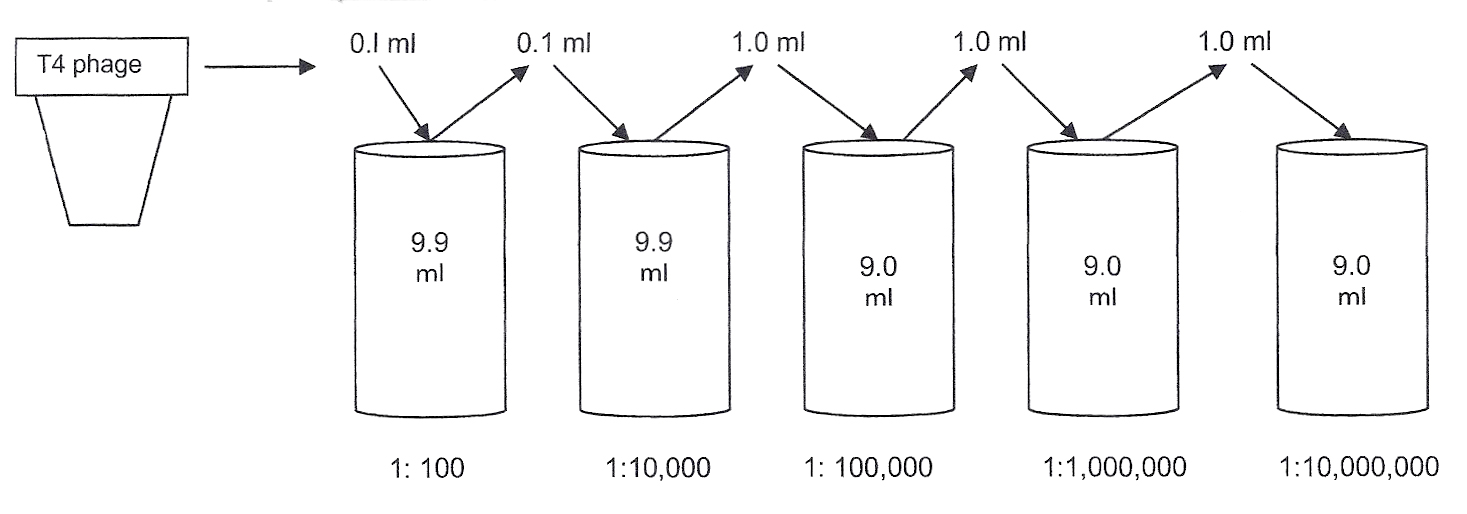
Diluting the original T4 phage sample:
Serial dilution of the T4 phage begins the quantitation procedure.
To calculate the dilution factor used for a sample, compare the amount of sample to the total volume of broth in the container.
0.1 ml of T4 phage sample to be tested
0.1 ml of sample + 9.9 ml of broth = 10 ml total volume in tube = 1:100
The sample added to the next tube is not the original sample. It is the original sample already diluted 1:100. Thus, the previous dilution is multiplied by the next dilution.
0.1 ml of diluted sample
0.1 ml of diluted sample+ 9.9 ml of broth = 1:10,000 dilution of the original T4 phage sample
The sample added to the next broth is taken from the previous dilution. Thus, this sample has already been diluted 10,000 times.
1 ml of the further diluted sample
1 ml of diluted sample + 9.0 ml broth = 1:100,000 dilution of the original T4 phage sample
You will continue diluting the original sample to obtain further dilutions of 1:1,000,000 and 1:10,000,000. These dilutions will help you find a dilution of virus in which 30-300 plaques can be counted.
Calculating the PFU:
The reason for performing the exercise is to count the T4 phages (PFU) in 1.0 ml of the original sample. To determine this, find the plate that has the countable number of PFU and count the plaques. This plaque number must be multiplied by the dilution (used to make that plate) and the amount of the sample taken.
PROCEDURE
Making serial dilutions of T4 Phage
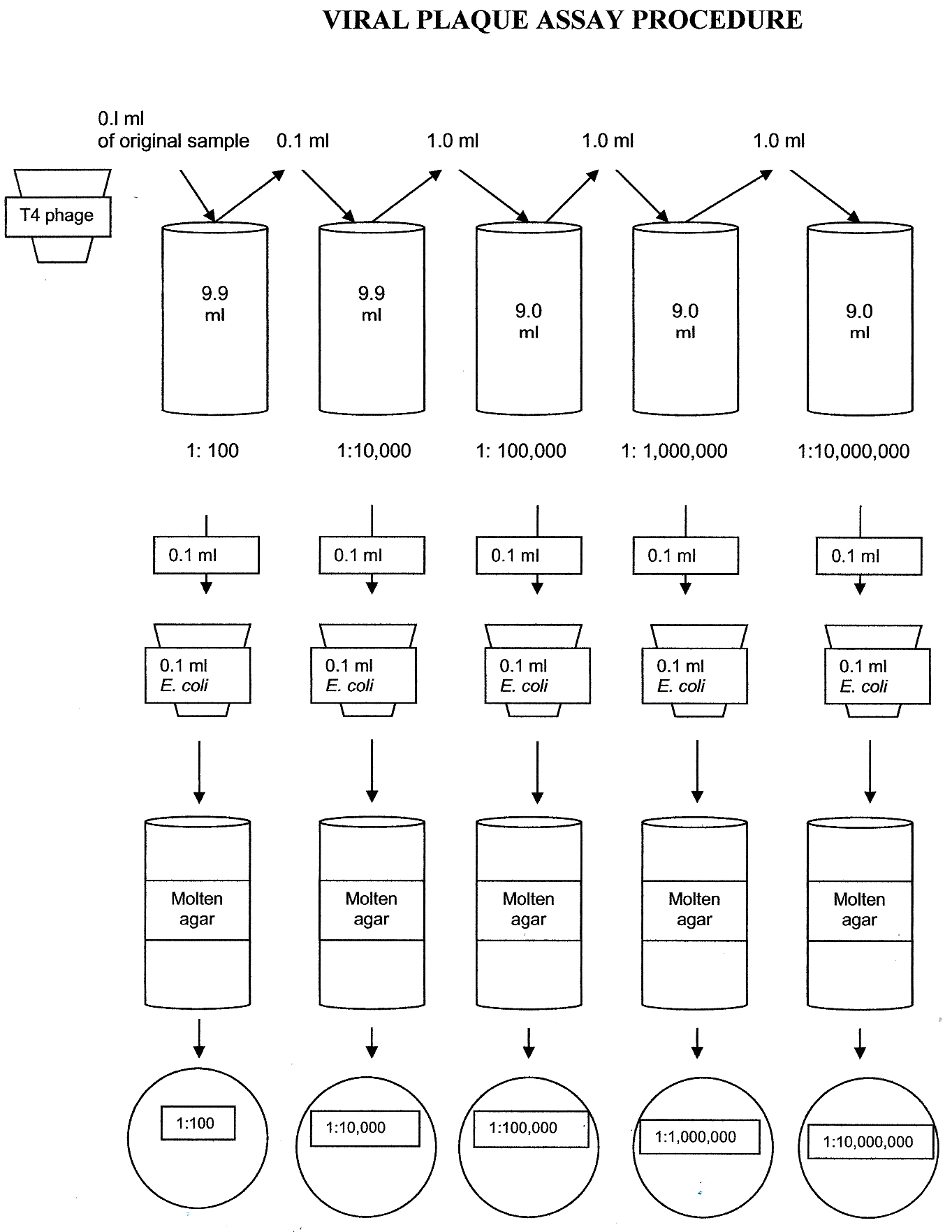
1. Obtain 2 tubes of Luria broth, each containing 9.9 ml. Also obtain 3 tubes of Luria broth, each containing 9.0 ml. Be careful to label the tubes correctly (make sure you keep the 9.9 and 9.0 tubes in order.) Label the tubes as 1:100, 1:10,000, 1:100,000, 1:1,000,000, 1:10,000,000
FOR EACH OF THE FOLLOWING DILUTIONS: Always use a new pipette tip and transfer pipette.
1. Transfer 0.1 ml bacteriophage into the luria broth tube labeled 1:100. Discard the tip in the autoclave trash. Mix the broth using a transfer pipette by taking in and expelling the fluid back into the tube several times. Discard the transfer pipette in the autoclave trash.
2. Transfer 0.1 ml of the 1:100 dilution into the luria broth labeled 1:10,000. Mix the broth using a transfer pipette by taking in and expelling the fluid back into the tube several times. Discard the transfer pipette in the autoclave trash.
3. Transfer 1.0 ml of the 1: 10,000 dilution into the luria broth tube labeled 1:100,000. Mix the broth using a transfer pipette by taking in and expelling the fluid back into the tube several times. Discard the transfer pipette in the autoclave trash.
4. Transfer 1.0 ml of the 1: 100,000 dilution into the luria broth tube labeled 1:1,000,000. Mix the broth using a transfer pipette by taking in and expelling the fluid back into the tube several times. Discard the transfer pipette in the autoclave trash.
5. Transfer 1.0 ml of the 1: 1,000,000 dilution into the luria broth tube labeled 1:10,000,000. Mix the broth using a transfer pipette by taking in and expelling the fluid back into the tube several times. Discard the transfer pipette in the autoclave trash.
Mix bacteriophage dilutions with E.coli
1. Label 5 microcentrifuge tubes the same way you labeled the original test tube dilutions – 1:100, 1:10,000, 1:100,000, 1:1,000,000, 1:10,000,000.
2. Pipette 0.1 ml E. coli into each of the microcentrifuge tubes using one pipette tip.
3. Using a different pipette tip for each transfer, pipette 0.1 ml of each phage dilution into the corresponding microcentrifuge tube of E. coli. (0.1 ml of 1:100 phage dilution into the tube of E. coli labeled 1:100, etc.) Rotate the tubes in your palms to mix them.
4. Incubate the microcentrifuge tubes containing the bacteriophage and E. coli at room temperature for 5 minutes.
Plating the phage/E.coli mixture
1. Obtain 5 Luria agar plates. Label the top side of each plate with your group name and the corresponding dilutions of the phage/E.coli mixtures. (1:100, 1:10,000, 1:100,000, 1:1,000,000, 1:10,000,000)
2. Obtain one tube of melted top agar from the water bath. (ONLY REMOVE ONE TUBE OF AGAR AT A TIME – IT SOLIDIFIES QUICKLY!).
3. Transfer the contents of the E. coli/phage mixture with a sterile transfer pipette into the melted top agar and gently mix. Immediately pour the contents onto the corresponding Luria agar plate. Quickly swirl the plate gently so that the liquid agar spreads evenly over the surface of Luria agar. Avoid producing bubbles which will affect the results.
4. Repeat Step 2 for each phage/E.coli dilution.
5. Incubate all plates right side up in the class plate tray at 37o C until the next lab session.
Calculating the number of bacteriophages in the original sample
1. Obtain the 5 agar plates and spread them out in order of dilution. Determine which dilution of bacteriophage produced between 30-300 plaques on the agar plate. Dispose of the other dilution plates in the autoclave trash.
2. Count the number of plaques on the agar plate and record the number on the worksheet.
3. Calculate the number of bacteriophage (Plaque Forming Units) in 1 ml of the original sample.
Plaque count X Plate Dilution x 10 (if you plated 0.1 ml)
You need to multiply by 10 if you plate 0.1 ml because we are calculating PFU/ml not PFU/0.1ml
Let’s do an example. You have performed a serial dilution of a sample with an unknown concentration of microorganisms. You counted 122 plaques on a plate inoculated with 0.1 ml of a 1:10,000 dilution. What was the population density of the original sample per milliliter (PFU/ml)?
122 X 10000 X 10 = 12,200,000 PFU/ml
To express 12,200,000 in scientific notation it needs to be a number between 1-9. The decimal point is at the end of the number, move the decimal to the left until you have a number between 1-9. The exponent will be positive.
1.22 x 107 PFU/ml
calculate the number of bacteriophages in the original sample-DEMONSTRATION PLATE SET
Below are photos of the five agar plates including the dilution and colony or plaque count. Determine which dilution of bacteriophage produced between 30-300 plaques (not colonies) on the agar plate. Calculate the number of bacteriophage (Plaque Forming Units) in 1 ml of the original sample using the data from the plate with 30-300 plaques.
Virus Plaques X Virus Dilution X 10 = PFU/1 ml sample
RECORD YOUR CALCULATION IN THE QUANTITATION OF MICROORGANISMS DEMONSTRATION QUESTIONS DOCUMENT.
Use this video to help you do the math.
DEMONSTRATION PLATE PHOTOS-SELECT THE PLATE BELOW WITH 30-300 PLAQUES (NOT COLONIES). CALCULATE PFU/ML AND RECORD THE CALCULATION ON THE QUESTIONS DOCUMENT.
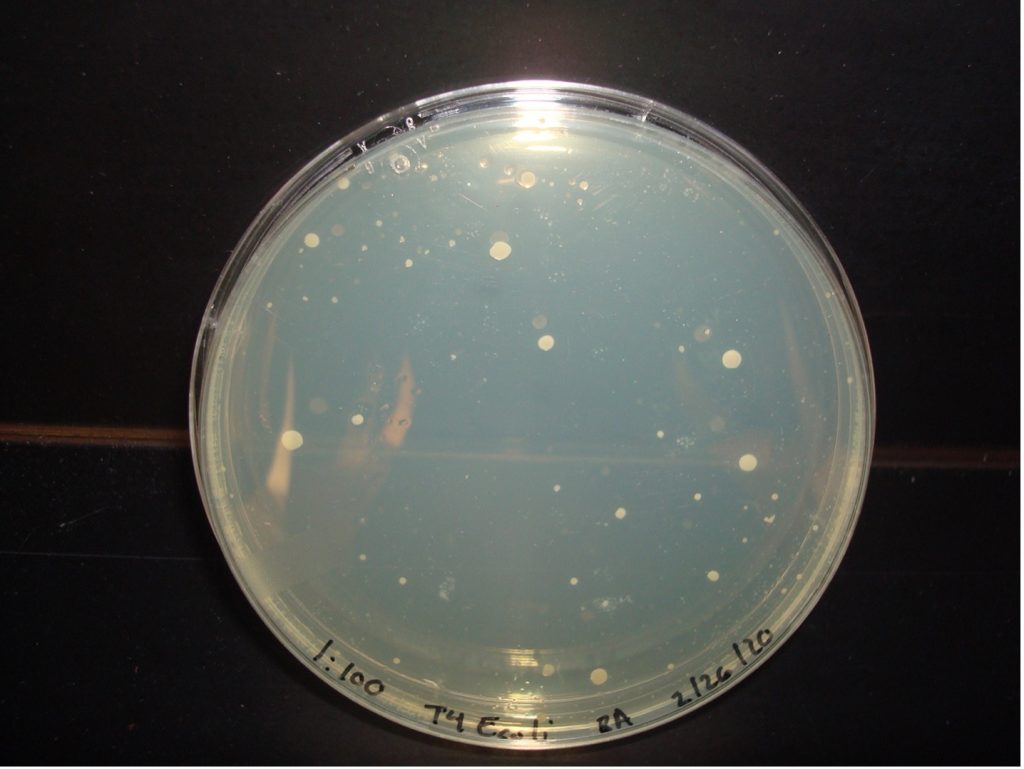
E. coli 1:100 Dilution 53 colonies
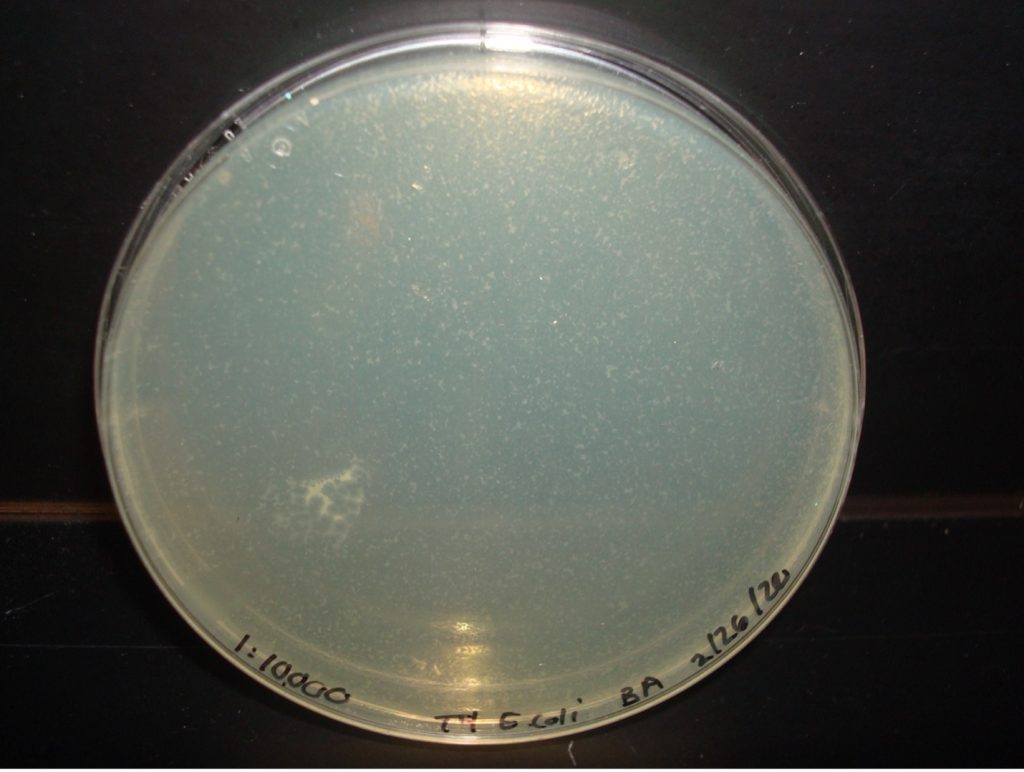
E. coli 1:10,000 Dilution too numerous to count plaques
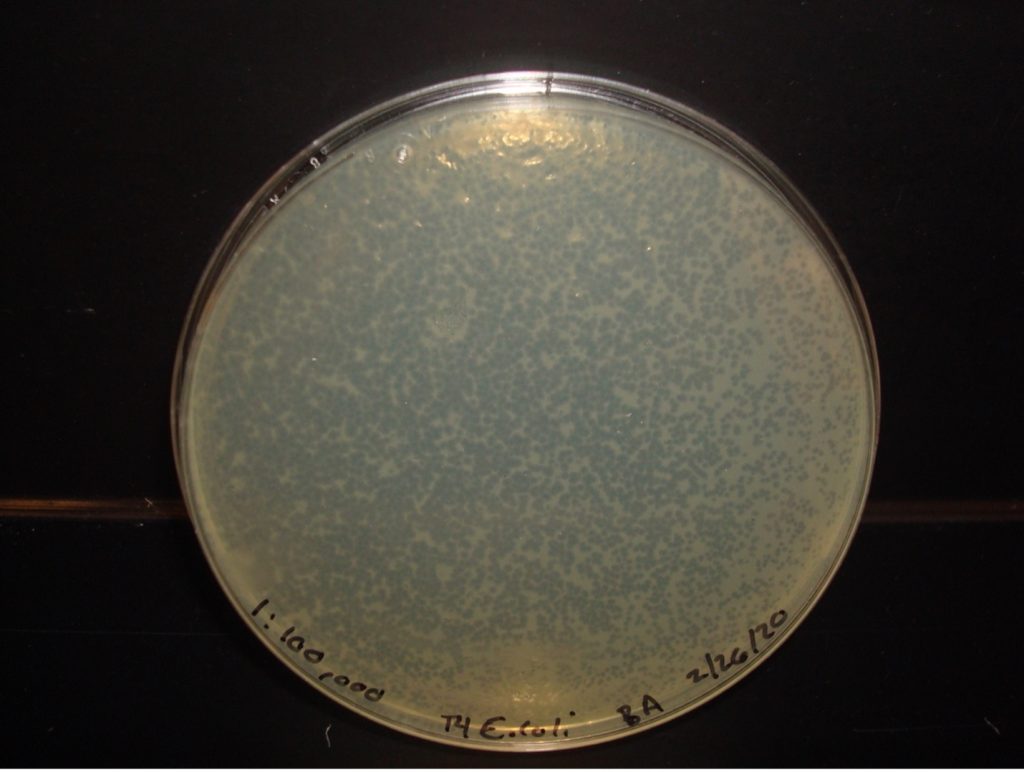
E. coli 1:100,000 Dilution too numerous to count plaques
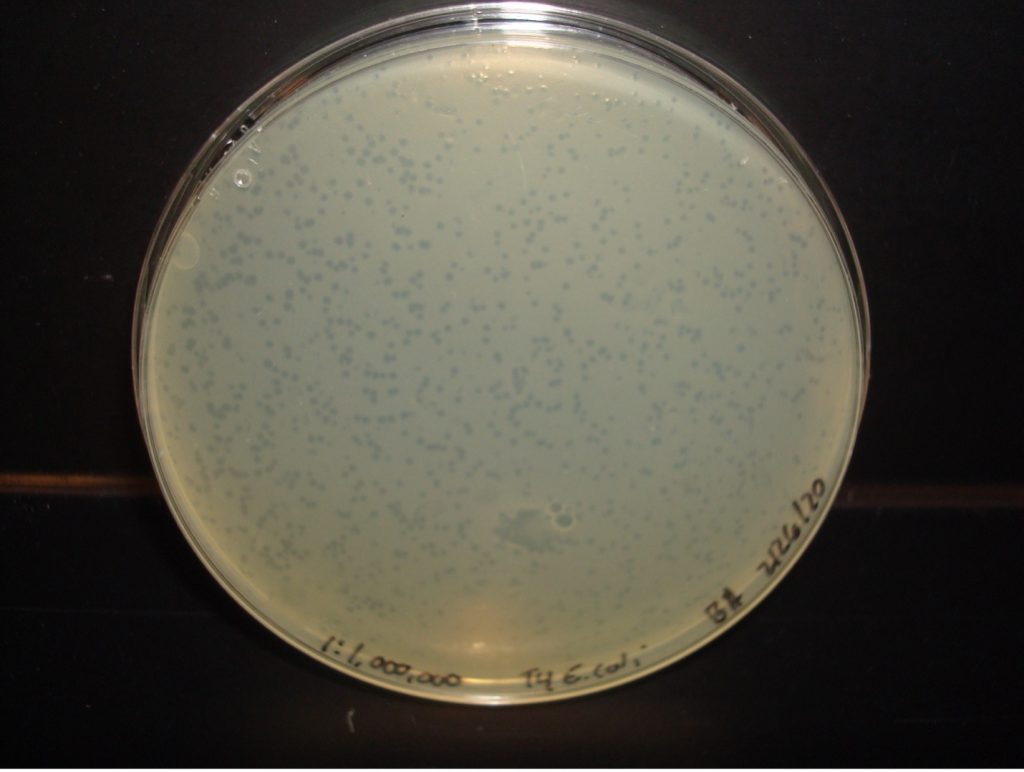
E. coli 1:1,000,000 Dilution too numerous to count plaques
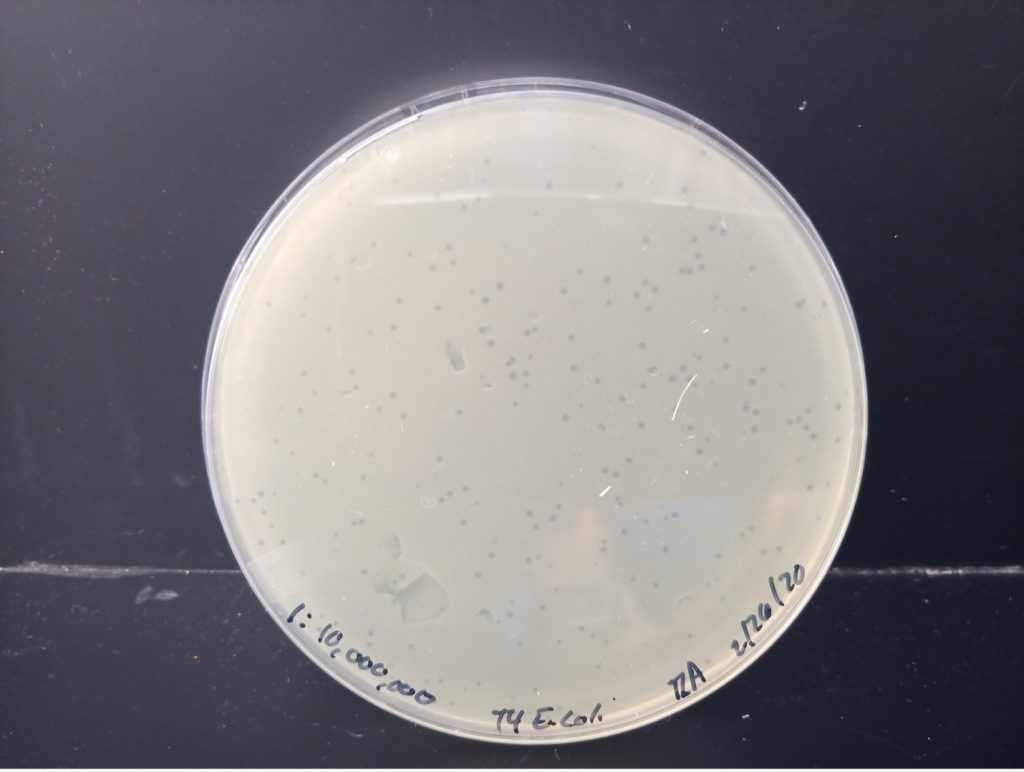
E. coli 1:10,000,000 Dilution 212 plaques
discoveries in microbiology

DR. LOUIS PASTEUR
French chemist and microbiologist, Dr. Pasteur developed a rabies vaccine that contained weakened rabies virus from a rabid rabbit. He injected a 60-year-old man who left the hospital after one injection. A 10-year-old girl was given one injection but died of rabies before another injection could be given. In 1885, 9-year-old Joseph Meister’s mother brought him to Pasteur after he was bitten by a rabid dog. Pasteur knew the boy would die from rabies if he did nothing. Pasteur injected Joseph with the vaccine every day for a total of thirteen days. Each successive injection contained less-weakened (stronger) virus. Meister never developed rabies, so the vaccination was a success. Later in life, Meister worked as caretaker of Pasteur’s tomb in Paris. The CDC recommends rabies vaccination for veterinarians, animal handlers, veterinary students, rabies laboratory workers, and spelunkers. With no previous exposure to rabies, the CDC recommends three doses of the rabies vaccine. With exposure to rabies, the CDC recommends four doses of the rabies vaccine and an injection of rabies immune globulin for an unvaccinated person, two doses of the rabies vaccine for a vaccinated person.
