TRANSFORMATION
LEARNING OBJECTIVES
Explain genetic transfer by transformation
Perform DNA recombination via transformation procedure
Observe altered bacterial characteristics due to transformation
MCCCD OFFICIAL COURSE COMPETENCIES
Describe the replication of genetic information, protein synthesis, and mutation in bacteria and viruses
Compare and contrast microbial methods of genetic recombination including transformation, conjugation, and transduction
Describe techniques and applications of genetic engineering and discuss their ethical implications
Describe modes of regulation of bacterial gene expression
Identify structural characteristics of the major groups of microorganism
Compare and contrast prokaryotic cell and eukaryotic cells
Compare and contrast the physiology and biochemistry of the various groups of microorganisms
Apply various laboratory techniques to identify types of microorganisms
Utilize aseptic technique for safe handling of microorganisms
MATERIALS
Materials: per table
Media:
1 Luria agar plate
2 Luria + Ampicillin agar plates
1 Luria + Ampicillin + Arabinose agar plate
1 tube of sterile Luria Broth
Equipment:
1 microcentrifuge tube rack
2 sterile microcentrifuge tubes
3 sterile disposable transfer pipettes
250 ml beaker of crushed ice
37oC water bath
Instructor’s desk:
pGLO plasmid (5 µL to each plasmid vial)
Escherichia coli 10-beta competent high efficiency (50 µL to each vial)
Micropipetters and sterile tips
BACTERIA ALBUM
There are two locations of DNA in a bacterial cell. Chromosomal DNA in a bacterial cell carries genes needed for the hereditary characteristics that are essential for bacterial growth and reproduction. Plasmids are small pieces of circular DNA that are separate from the chromosome and replicate independently. Bacteria naturally have one or more plasmids with genes for one or more traits that may be beneficial, but not essential, to bacterial survival. In nature, bacteria can transfer plasmids back and forth allowing them to share these beneficial genes. This natural mechanism allows bacteria to adapt to new environments. The recent occurrence of bacterial resistance to antibiotics is due to the transmission of plasmids.
A gene is a segment of DNA which “codes for” or provides the instructions for making a protein. This protein gives an organism a trait. Genes can be transferred from one organism to another by transformation which involves the insertion of the gene into an organism to change the organism’s trait.
DNA –> RNA –> Protein –> Trait
The transfer of genes by the transformation process is used in many areas of biotechnology. In agriculture, genes coding for traits such as frost, pest, or spoilage resistance can be genetically transformed into plants. In bioremediation, bacteria can be genetically transformed with genes enabling them to digest oil spills. In medicine, diseases caused by defective genes are treated by gene therapy; that is, by genetically transforming a sick person’s cells with healthy copies of the defective gene that causes the disease. The GFP gene in this exercise was originally found in the bioluminescent jellyfish Aequorea Victoria. The GFP gene codes for green fluorescent protein which allows the jellyfish to fluoresce and glow in the dark.
This procedure transforms E. coli bacteria by introducing a unique, genetically engineered pGLO plasmid in which three genes have been inserted. Following the transformation procedure, the competent bacteria cells (able to take up the plasmid) will express their newly acquired genes. Most bacterial cells are not competent; however, mixing the cells with calcium chloride during their growth phase will induce competence by the interactions of calcium ions with the cell envelope and the DNA phosphate backbone.
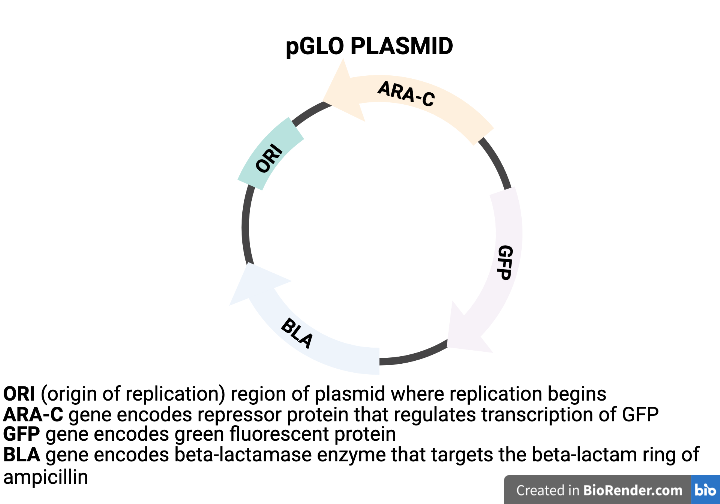
The three genes inserted on the pGLO plasmid are listed in the table. Each gene codes for a specific protein which will enable the E. coli to demonstrate different traits.
| GENE | PROTEIN | TRAIT |
| BLA | Beta-lactamase | Breaks apart ampicillin (targets beta-lactam ring) |
| Ara-C | Repressor | Regulates transcription of GFP gene |
| GFP | Green Fluorescent protein | Glows under UV light |
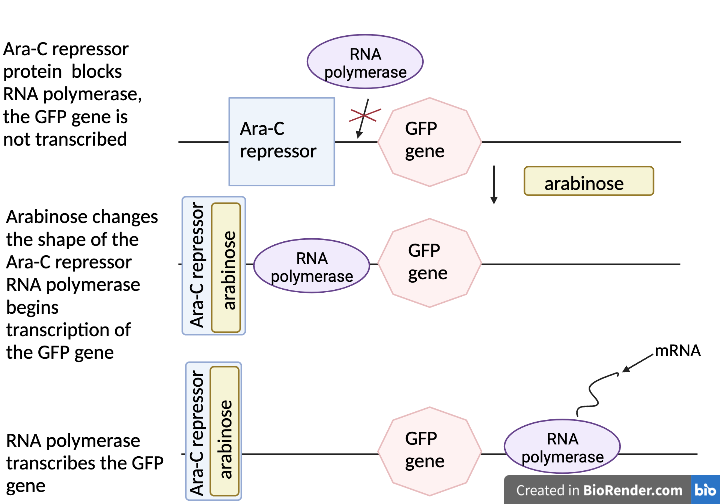
pGLO incorporates an inducible operon which controls the expression of the Fluorescent Protein in transformed cells. The BLA gene and Ara-C gene are continually transcribed and translated into protein. The gene for GFP, on the other hand, is regulated by the Repressor protein and will not be transcribed without the addition the sugar arabinose to the cells’ nutrient medium. The arabinose will inactivate the repressor protein thus allowing the GFP gene to be transcribed.
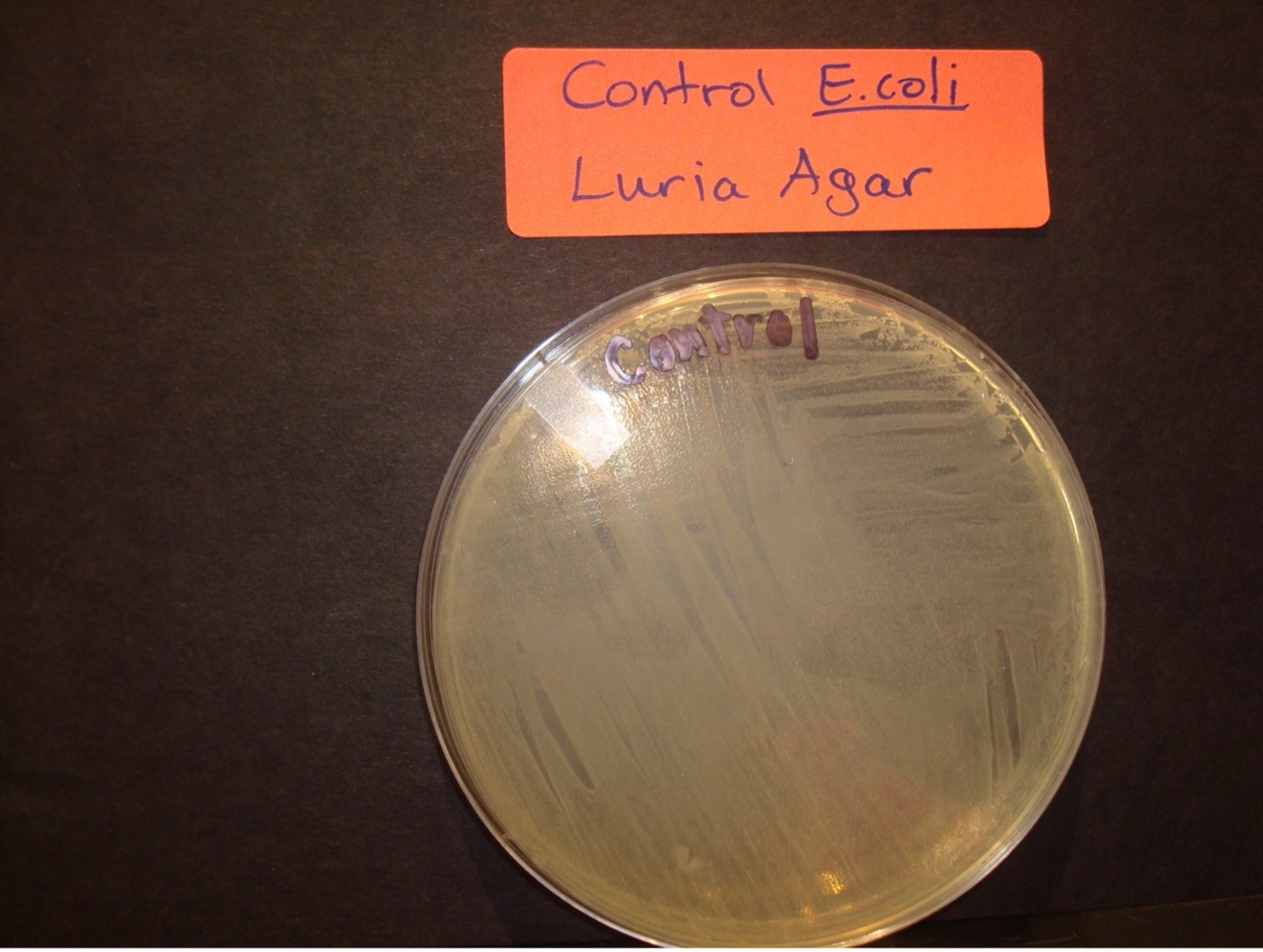
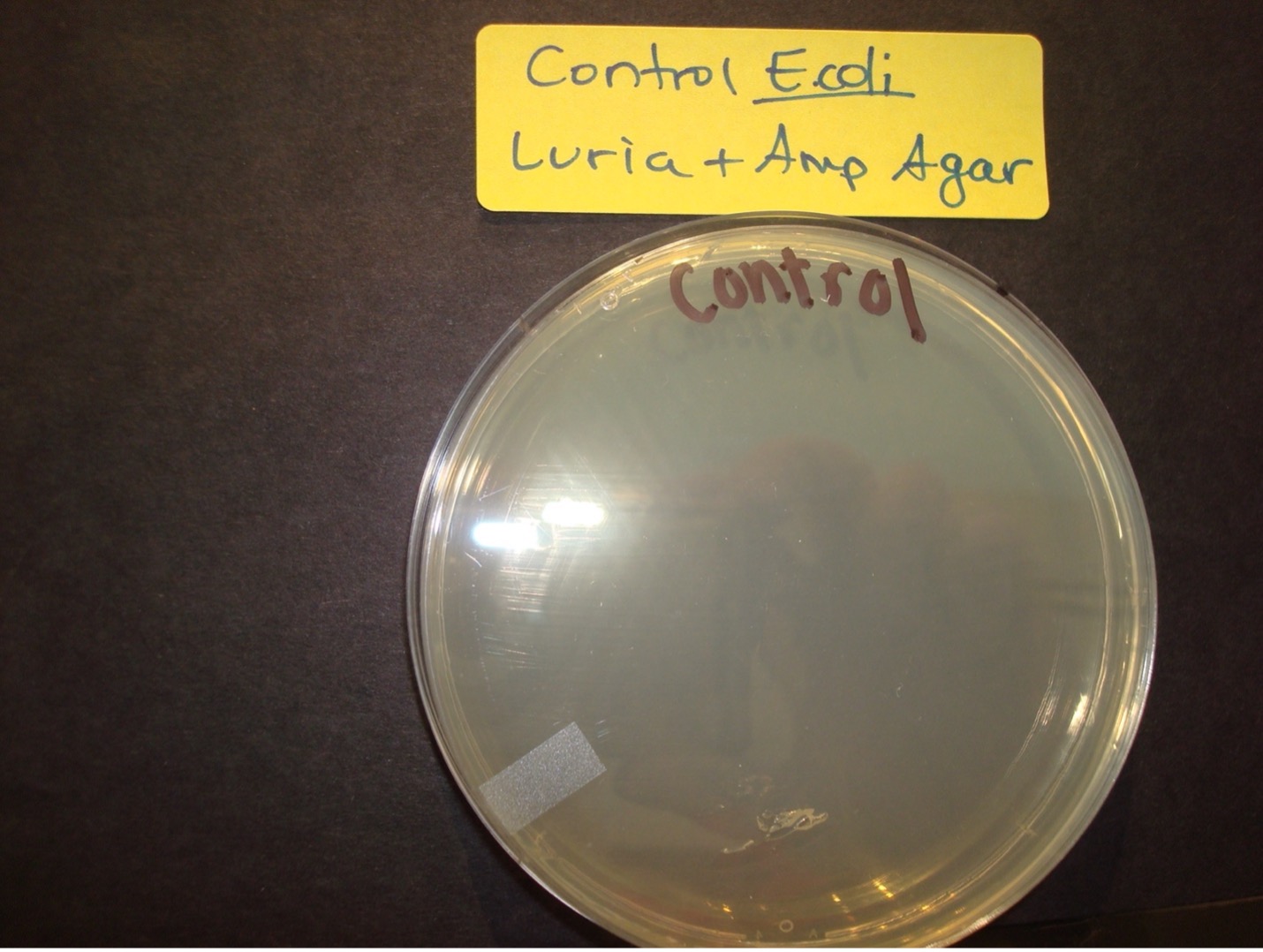
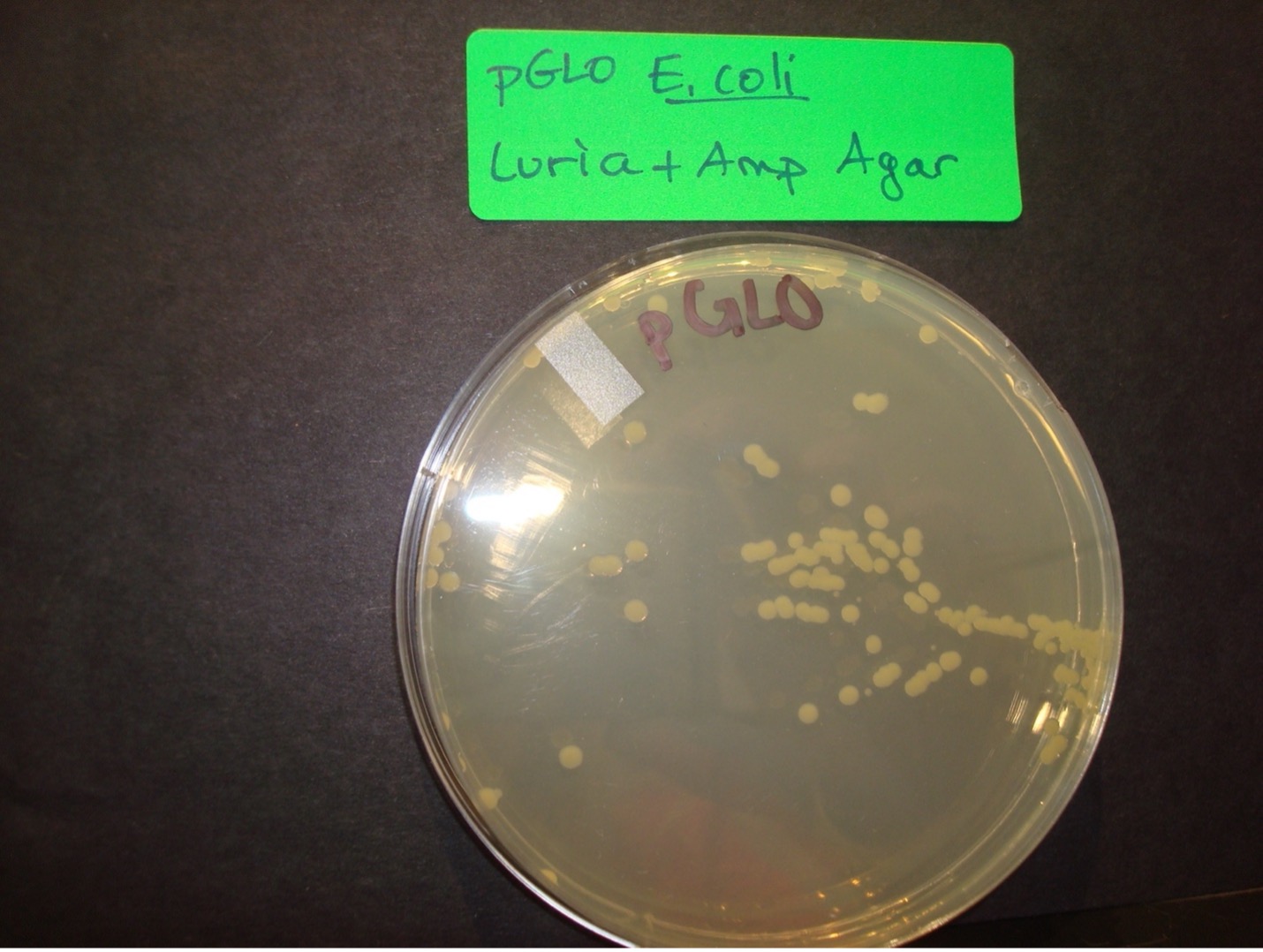
Selection for E. coli that are transformed with pGLO is accomplished by growth on nutrient agar containing the beta-lactam antibiotic, ampicillin, and on nutrient agar containing ampicillin plus the sugar, arabinose. Transformed cells will demonstrate the traits of the additional genes. The E. coli that were not transformed will be killed by the antibiotic, ampicillin.
Pre-Assessment
PROCEDURE
Transformation Procedure
1. Obtain a beaker of ice and two microcentrifuge tubes. Label the lid of one tube “P” (pGLO + E. coli) and the other lid “C” for control (only E. coli).
2. Take both tubes to the instructor, who will pipette 5µL pGLO plasmid into the tube marked “P” and 50µL E. coli into both tubes.
3. Immediately place both tubes in the beaker of ice for 20 minutes.
4. Transfer the tubes into the 37oC water bath for EXACTLY 3 minutes. During this time, the competent E. coli are taking up the plasmid. The heat shock alters membrane fluidity creating pores and allows for plasmid DNA to enter the bacterial cell.
5. Immediately transfer the tubes back into the ice for one minute.
6. Add 0.5mL Luria broth to each tube and incubate in the 37oC water bath for at least 30 minutes. The transformed E. coli use the nutrients and time to “express” the genes.
7. While the E. coli is incubating, label the following media with your group name:
1 Luria agar plate with Ampicillin and Arabinose – Label “pGLO” E. coli
2 Luria agar plates with Ampicillin – Label one with “pGLO” E. coli, Label one with “Control” E. coli
1 plain Luria agar plate – Label “Control” E. coli
8. After the 30 minute incubation period, use a sterile transfer pipette to transfer half of the pGLO E. coli mixture onto the Luria agar with Ampicillin and arabinose pGLO plate and half of the mixture onto the Luria agar with Ampicillin pGLO E. coli plate.
9. Use a sterile spreader to spread the liquid over the surface of both of the pGLO E. coli agar plates. Dispose of the “P” microcentrifuge tube and the spreader in the autoclave trash.
10. Use another sterile transfer pipette to transfer half of the “Control” E. coli mixture to the Luria agar with Ampicillin and half of the mixture to the plain Luria agar.
11. Use a sterile spreader to spread the liquid over the surface of both of the control E. coli agar plates. Dispose of the “C” microcentrifuge tube and spreader in the autoclave trash.
12. Incubate the inverted plates in the class plate tray until the next lab session.
AFTER INCUBATION-Record the results in the worksheet.

POST TEST
DISCOVERIES IN MICROBIOLOGY
 DR. ÉLIE METCHNIKOFF
DR. ÉLIE METCHNIKOFF
In 1882, Russian zoologist and microbiologist Dr. Metchinkoff was the first to discover phagocytosis and phagocytes. He advocated the use of lactic acid bacteria (Lactobacillus) for healthy and long life. This became the concept of probiotics in medicine.

